Standard Interpolation#
Interpolate 2D images with standard interpolation. Compare them to SciPy zoom. We compute SNR and MSE on a face ROI (not the whole frame) to focus on detail and avoid boundary artifacts.
Imports#
import numpy as np
import time
from urllib.request import urlopen
from PIL import Image
from scipy.ndimage import zoom as _scipy_zoom
from splineops.resize import resize
from splineops.utils.metrics import compute_snr_and_mse_region
from splineops.utils.plotting import plot_difference_image, show_roi_zoom
from splineops.utils.diagram import draw_standard_vs_scipy_pipeline
def fmt_ms(seconds: float) -> str:
"""Format seconds as a short 'X.X ms' string."""
return f"{seconds * 1000.0:.1f} ms"
# You can switch this to np.float64 if you want full double precision.
DTYPE = np.float32
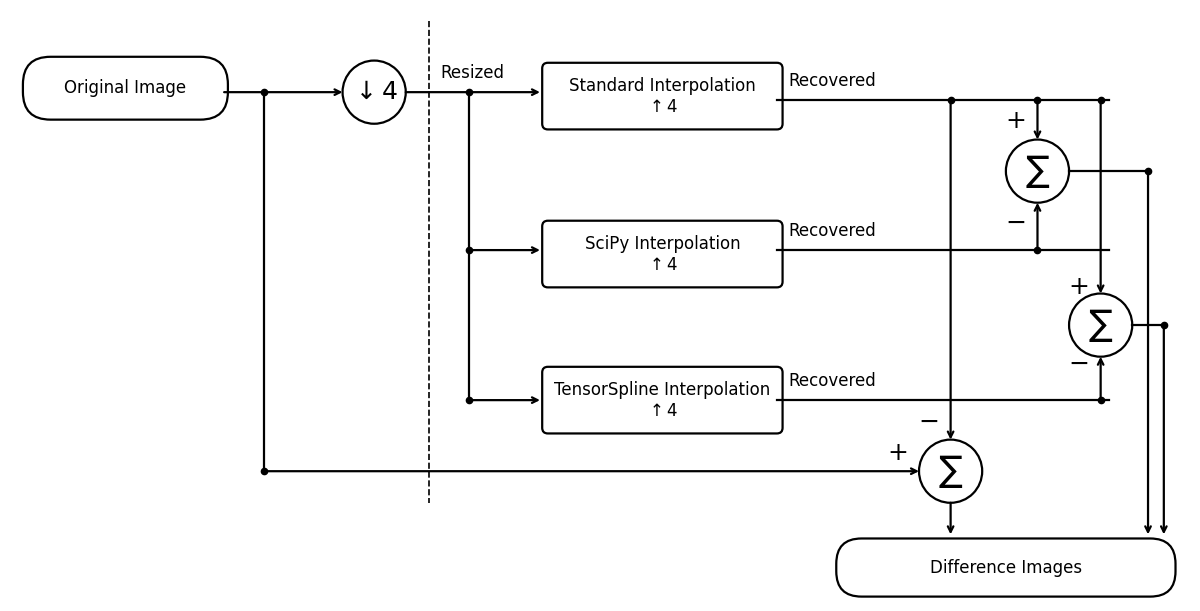
Pipeline Diagram#
These experiments validate the standard interpolation against SciPy’s by showing they produce (nearly) the same result, and where tiny differences are.
Load and Normalize an Image#
Here, we load an example image from an online repository and convert to grayscale in [0, 1].
url = 'https://r0k.us/graphics/kodak/kodak/kodim14.png'
with urlopen(url, timeout=10) as resp:
img = Image.open(resp)
# Start in float64 for robust normalization, then cast once to DTYPE.
data = np.array(img, dtype=np.float64)
# Convert to [0..1]
input_image_normalized = data / 255.0
# Convert to grayscale via simple weighting
input_image_normalized = (
input_image_normalized[:, :, 0] * 0.2989 + # Red channel
input_image_normalized[:, :, 1] * 0.5870 + # Green channel
input_image_normalized[:, :, 2] * 0.1140 # Blue channel
)
# Run the interpolation backends in DTYPE (e.g. float32 for speed).
input_image_normalized = input_image_normalized.astype(DTYPE, copy=False)
zoom = np.pi / 6 # ≈ 0.5235987756
zoom_factors_2d = (zoom, zoom)
border_fraction = 0.3 # still available as a fallback (unused when roi=... is set)
# Face-centered 64×64 ROI (focus region for metrics & diffs)
ROI_SIZE_PX = 64
FACE_ROW, FACE_COL = 400, 600 # (row, col) approx center of the detail
h_img, w_img = input_image_normalized.shape
# Top-left of the 64×64 box, clipped to stay inside the image
row_top = int(np.clip(FACE_ROW - ROI_SIZE_PX // 2, 0, h_img - ROI_SIZE_PX))
col_left = int(np.clip(FACE_COL - ROI_SIZE_PX // 2, 0, w_img - ROI_SIZE_PX))
# ROI rectangle for metrics/plots (row, col, height, width)
roi_rect = (row_top, col_left, ROI_SIZE_PX, ROI_SIZE_PX)
roi_kwargs = dict(
roi_height_frac=ROI_SIZE_PX / h_img, # keeps height at 64 px (square ROI)
grayscale=True,
roi_xy=(row_top, col_left), # top-left of the ROI
)
# Original (shifted ROI)
_ = show_roi_zoom(
input_image_normalized,
ax_titles=("Original Image", None),
**roi_kwargs
)

Standard Interpolation#
We use our standard interpolation method (cubic). SNR/MSE are computed on the face ROI.
# Forward + backward resize with splineops.resize.resize
t0 = time.perf_counter()
resized_2d_interp = resize(
input_image_normalized,
zoom_factors=zoom_factors_2d,
method="cubic",
)
t1 = time.perf_counter()
recovered_2d_interp = resize(
resized_2d_interp,
output_size=input_image_normalized.shape,
method="cubic",
)
t2 = time.perf_counter()
time_2d_interp_fwd = t1 - t0
time_2d_interp_back = t2 - t1
time_2d_interp = t2 - t0 # total pipeline time
# Metrics (ROI-aware)
snr_2d_interp, mse_2d_interp = compute_snr_and_mse_region(
input_image_normalized,
recovered_2d_interp,
roi=roi_rect,
border_fraction=border_fraction,
)
Resized Image#
Show the resized image pasted on a white canvas (for zoom-out), plus the ROI zoom.
# Zoomed face detail for the resized (cubic) image pasted onto original-size canvas
h_res, w_res = resized_2d_interp.shape
zoom_r, zoom_c = zoom_factors_2d
# ROI size in the resized image (e.g., 64 -> 16 px when zoom=0.25)
roi_h_res = max(1, int(round(ROI_SIZE_PX * zoom_r)))
roi_w_res = max(1, int(round(ROI_SIZE_PX * zoom_c)))
# ROI center mapped into the resized image
center_r_res = int(round(FACE_ROW * zoom_r))
center_c_res = int(round(FACE_COL * zoom_c))
# Top-left of the ROI in the resized image, clipped to bounds
row_top_res = int(np.clip(center_r_res - roi_h_res // 2, 0, h_res - roi_h_res))
col_left_res = int(np.clip(center_c_res - roi_w_res // 2, 0, w_res - roi_w_res))
# --- Build original-size white canvas and paste the small resized image at top-left (0,0) ---
canvas = np.ones((h_img, w_img), dtype=resized_2d_interp.dtype) # white background in [0,1]
canvas[:h_res, :w_res] = resized_2d_interp
roi_kwargs_on_canvas = dict(
roi_height_frac=roi_h_res / h_img, # keeps the inset square at roi_h_res pixels high
grayscale=True,
roi_xy=(row_top_res, col_left_res), # same coords since pasted at (0,0)
)
_ = show_roi_zoom(
canvas,
ax_titles=(f"Resized Image (standard, {fmt_ms(time_2d_interp_fwd)})", None),
**roi_kwargs_on_canvas
)

Recovered Image (Standard)#
We plot the recovered image (after reversing the zoom) with the same ROI.
_ = show_roi_zoom(
recovered_2d_interp,
ax_titles=(f"Recovered Image (standard, {fmt_ms(time_2d_interp_back)})", None),
**roi_kwargs
)

SciPy Interpolation#
For comparison, we also use SciPy’s zoom method. Metrics are computed on the ROI. SciPy preserves the input dtype, so it will also run in DTYPE here.
t0 = time.perf_counter()
resized_2d_scipy = _scipy_zoom(
input_image_normalized,
zoom_factors_2d,
order=3,
)
t1 = time.perf_counter()
recovered_2d_scipy = _scipy_zoom(
resized_2d_scipy,
1.0 / np.asarray(zoom_factors_2d),
order=3,
)
t2 = time.perf_counter()
time_2d_scipy_fwd = t1 - t0
time_2d_scipy_back = t2 - t1
time_2d_scipy = t2 - t0 # total pipeline time
snr_2d_scipy, mse_2d_scipy = compute_snr_and_mse_region(
input_image_normalized,
recovered_2d_scipy,
roi=roi_rect,
border_fraction=border_fraction,
)
Recovered Image (SciPy)#
_ = show_roi_zoom(
recovered_2d_scipy,
ax_titles=(f"Recovered Image (SciPy, {fmt_ms(time_2d_scipy_back)})", None),
**roi_kwargs
)

Difference Images#
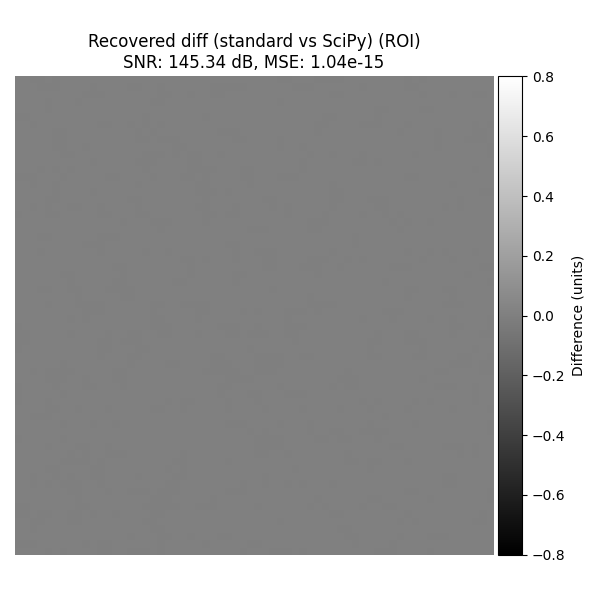
Standard vs SciPy#
Compare the two recovered images on the face ROI and plot that difference.
snr_scipy_vs_interp, mse_scipy_vs_interp = compute_snr_and_mse_region(
recovered_2d_scipy, recovered_2d_interp, roi=roi_rect
)
plot_difference_image(
original=recovered_2d_scipy,
recovered=recovered_2d_interp,
snr=snr_scipy_vs_interp,
mse=mse_scipy_vs_interp,
roi=roi_rect,
title_prefix="Recovered diff (standard vs SciPy)",
)
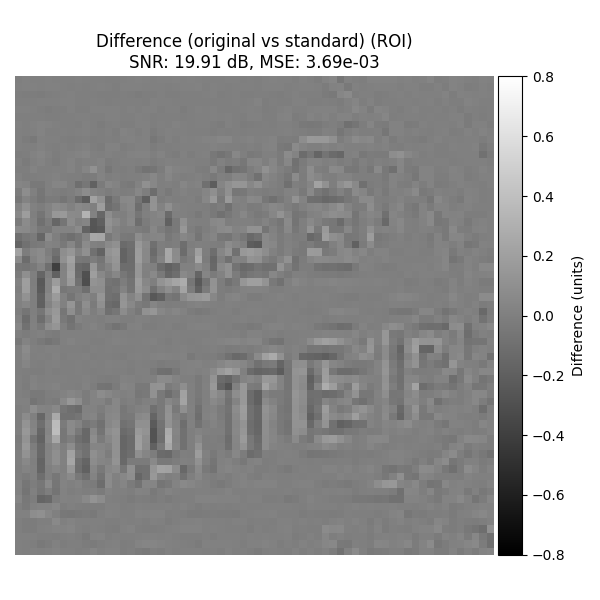
Original vs Standard#
For completeness, show the difference image (original - recovered with standard interpolation) on the ROI.
plot_difference_image(
original=input_image_normalized,
recovered=recovered_2d_interp,
snr=snr_2d_interp,
mse=mse_2d_interp,
roi=roi_rect,
title_prefix="Difference (original vs standard)",
)
Alternative using TensorSpline#
As an alternative, we can replicate the same interpolation manually using the
TensorSpline class, which underpins the resize() function behind the scenes.
from splineops.spline_interpolation.tensor_spline import TensorSpline
# 1) Build uniform coordinate arrays that match the shape of 'input_image_normalized'
height, width = input_image_normalized.shape
# Use the same dtype as the image for coordinates, so everything lives in DTYPE.
x_coords = np.linspace(0, height - 1, height, dtype=input_image_normalized.dtype)
y_coords = np.linspace(0, width - 1, width, dtype=input_image_normalized.dtype)
coordinates_2d = (x_coords, y_coords)
# 2) For "cubic interpolation", pick "bspline3".
# For boundary handling, we can pick "mirror", "zero", etc.
ts = TensorSpline(
data=input_image_normalized,
coordinates=coordinates_2d,
bases="bspline3", # cubic B-splines
modes="mirror" # handles boundaries with mirroring
)
# 3) Define new coordinate grids for the "zoomed" shape.
zoomed_height = int(height * zoom_factors_2d[0])
zoomed_width = int(width * zoom_factors_2d[1])
x_coords_zoomed = np.linspace(
0, height - 1, zoomed_height, dtype=input_image_normalized.dtype
)
y_coords_zoomed = np.linspace(
0, width - 1, zoomed_width, dtype=input_image_normalized.dtype
)
coords_zoomed_2d = (x_coords_zoomed, y_coords_zoomed)
# Evaluate (forward pass): zoom in or out
resized_direct_ts = ts(coordinates=coords_zoomed_2d)
# 4) Define coordinate grids for returning to the original shape
x_coords_orig = np.linspace(
0, height - 1, height, dtype=input_image_normalized.dtype
)
y_coords_orig = np.linspace(
0, width - 1, width, dtype=input_image_normalized.dtype
)
coords_orig_2d = (x_coords_orig, y_coords_orig)
# Evaluate (backward pass): from zoomed shape back to original
ts_zoomed = TensorSpline(
data=resized_direct_ts,
coordinates=coords_zoomed_2d,
bases="bspline3",
modes="mirror"
)
recovered_direct_ts = ts_zoomed(coordinates=coords_orig_2d)
# Now, resized_direct_ts / recovered_direct_ts should be very similar
# to 'resized_2d_interp' / 'recovered_2d_interp' from the high-level "resize()" approach.
# Let's compute MSE to confirm:
mse_forward = np.mean((resized_direct_ts - resized_2d_interp ) ** 2)
mse_backward = np.mean((recovered_direct_ts - recovered_2d_interp) ** 2)
print(f"MSE (TensorSpline vs. resize()) resized: {mse_forward:.6e}")
print(f"MSE (TensorSpline vs. resize()) recovered: {mse_backward:.6e}")
MSE (TensorSpline vs. resize()) resized: 7.308340e-13
MSE (TensorSpline vs. resize()) recovered: 1.133354e-12
Total running time of the script: (0 minutes 2.799 seconds)
