Wavelet Decomposition#
This example demonstrates how to use the Decompose module for Haar wavelet decomposition (analysis & synthesis) in 2D.
Imports#
import numpy as np
import matplotlib.pyplot as plt
# For downloading and handling the image
import requests
from io import BytesIO
from PIL import Image
# Wavelet classes for 2D
from splineops.decompose.wavelets.haar import HaarWavelets
from splineops.decompose.wavelets.splinewavelets import (
Spline1Wavelets,
Spline3Wavelets,
Spline5Wavelets
)
Load and Normalize a 2D Image#
Here, we load an example image from an online repository. We convert it to grayscale in [0,1].
url = 'https://r0k.us/graphics/kodak/kodak/kodim07.png'
response = requests.get(url)
img = Image.open(BytesIO(response.content))
# Convert to numpy float64
image_color = np.array(img, dtype=np.float64)
# Normalize to [0,1]
image_color /= 255.0
# Convert to grayscale using standard weights
image_gray = (
image_color[:, :, 0] * 0.2989 +
image_color[:, :, 1] * 0.5870 +
image_color[:, :, 2] * 0.1140
)
ny, nx = image_gray.shape
print(f"Downloaded image shape = {ny} x {nx}")
def imshow_matched_LL(
coeffs,
levels,
orig_image, # full-resolution grayscale image
detail_pct=95, # percentile for LH/HL/HH stretch
ll_low=2, ll_high=98,# LL stretch percentiles
ax=None,
title=None,
cmap='gray',
):
"""
Visualise a wavelet pyramid so that
• the smallest LL block has roughly the same contrast and overall
brightness as the original image; and
• every detail coefficient is stretched, then mapped to 0.5 ± 0.5 so
zero is mid-gray.
Parameters
----------
coeffs : 2-D np.ndarray
Wavelet-coefficient array in quadrant-pyramid layout.
levels : int
Decomposition depth (to locate the LL block).
orig_image : 2-D np.ndarray
Original full-resolution grayscale image in [0, 1].
detail_pct : float
|coeff| percentile that maps to ±1 in the detail bands.
ll_low, ll_high : float
Percentiles (0–100) used for LL contrast stretch.
"""
if ax is None:
ax = plt.gca()
vis = np.empty_like(coeffs, dtype=np.float64)
ny, nx = vis.shape
ny_ll = ny // (2 ** levels)
nx_ll = nx // (2 ** levels)
# ─────────────────── 1. LL block ───────────────────
ll = coeffs[:ny_ll, :nx_ll]
lo, hi = np.percentile(ll, [ll_low, ll_high])
hi = max(hi, lo + 1e-12) # avoid zero division
ll_lin = np.clip((ll - lo) / (hi - lo), 0, 1)
# ▸ match mean brightness to original image
mean_orig = float(np.mean(orig_image))
mean_ll = float(np.mean(ll_lin))
if mean_ll < 1e-12: # degenerate (all black): avoid log(0)
gamma = 1.0
else:
gamma = np.log(mean_orig + 1e-12) / np.log(mean_ll + 1e-12)
ll_matched = ll_lin ** gamma
vis[:ny_ll, :nx_ll] = ll_matched
# ─────────────────── 2. Detail bands ───────────────
detail_mask = np.ones_like(coeffs, dtype=bool)
detail_mask[:ny_ll, :nx_ll] = False
if detail_mask.any():
dvals = coeffs[detail_mask]
d_scale = np.percentile(np.abs(dvals), detail_pct)
d_scale = max(d_scale, 1e-12)
d_norm = np.clip(dvals / d_scale, -1, 1) / 2 + 0.5 # [-1,1]→[0,1]
vis[detail_mask] = d_norm
# ─────────────────── 3. Display ────────────────────
ax.imshow(vis, cmap=cmap, vmin=0, vmax=1, interpolation='nearest')
ax.axis('off')
if title:
ax.set_title(title, fontsize=14)
Downloaded image shape = 512 x 768
2D Wavelet Decomposition#
We demonstrate wavelet decomposition (analysis) and reconstruction (synthesis) using 2D Haar wavelets on a grayscale image.
haar2d = HaarWavelets(scales=3)
coeffs = haar2d.analysis(image_gray)
recon_haar = haar2d.synthesis(coeffs)
err_haar = recon_haar - image_gray
max_err_haar = np.abs(err_haar).max()
print("[Wavelets 2D Haar Test]")
print(f"Max error after 3-scale decomposition: {max_err_haar}")
# Helper function for visualization
def pyramid_with_quadrant_embedding_levels(wavelet, inp, num_levels):
"""
Perform multi-scale wavelet analysis in-place so that at each level the
new coarse approximation is stored in the quadrant corresponding to the
previous level's coarse region.
Parameters
----------
wavelet : AbstractWavelets instance
A wavelet instance (e.g., HaarWavelets) with the desired number of scales.
inp : np.ndarray
Input 2D array (e.g., grayscale image).
num_levels : int
The number of decomposition levels to perform.
Returns
-------
coeffs : np.ndarray
Final coefficient array (same size as inp) with the pyramid layout.
"""
out = np.copy(inp)
ny, nx = out.shape[:2]
for level in range(num_levels):
# Process the current top-left subarray
sub = out[:ny, :nx]
sub_out = wavelet.analysis1(sub)
out[:ny, :nx] = sub_out
# Update region size for next level (halve each dimension)
nx = max(1, nx // 2)
ny = max(1, ny // 2)
return out
[Wavelets 2D Haar Test]
Max error after 3-scale decomposition: 1.3322676295501878e-15
1-Level Decomposition#
wavelet1 = HaarWavelets(scales=1)
coeffs1 = pyramid_with_quadrant_embedding_levels(wavelet1, image_gray, 1)
plt.figure(figsize=(8, 8))
imshow_matched_LL(coeffs1, levels=1, orig_image=image_gray,
detail_pct=95, ll_low=5, ll_high=99,
title="Haar 1-Level Decomposition")
plt.tight_layout()
plt.show()

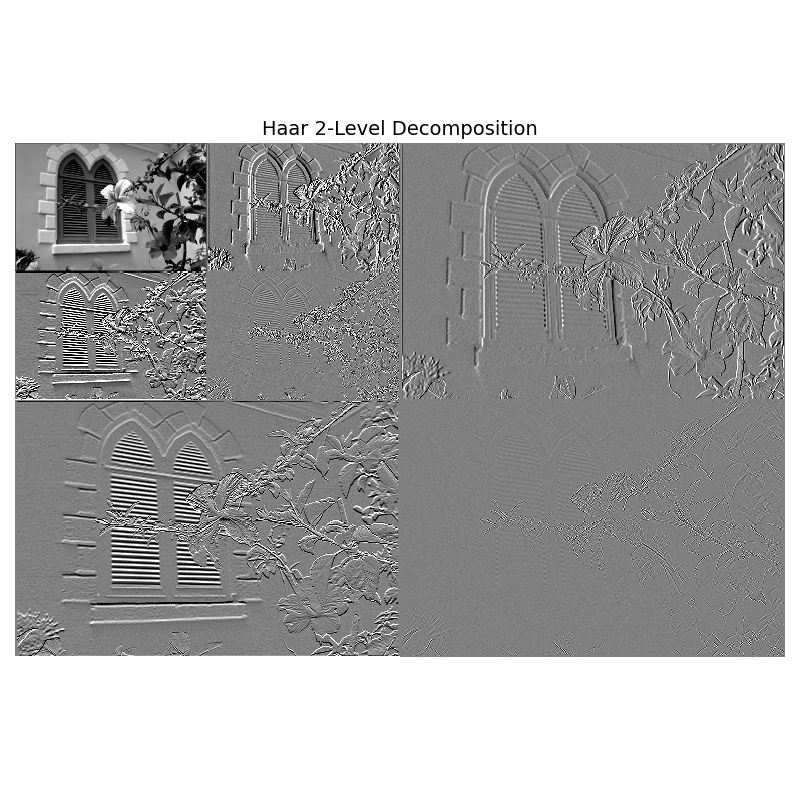
2-Level Decomposition#
wavelet2 = HaarWavelets(scales=2)
coeffs2 = pyramid_with_quadrant_embedding_levels(wavelet2, image_gray, 2)
plt.figure(figsize=(8, 8))
imshow_matched_LL(coeffs2, levels=2, orig_image=image_gray,
detail_pct=95, ll_low=5, ll_high=99,
title="Haar 2-Level Decomposition")
plt.tight_layout()
plt.show()
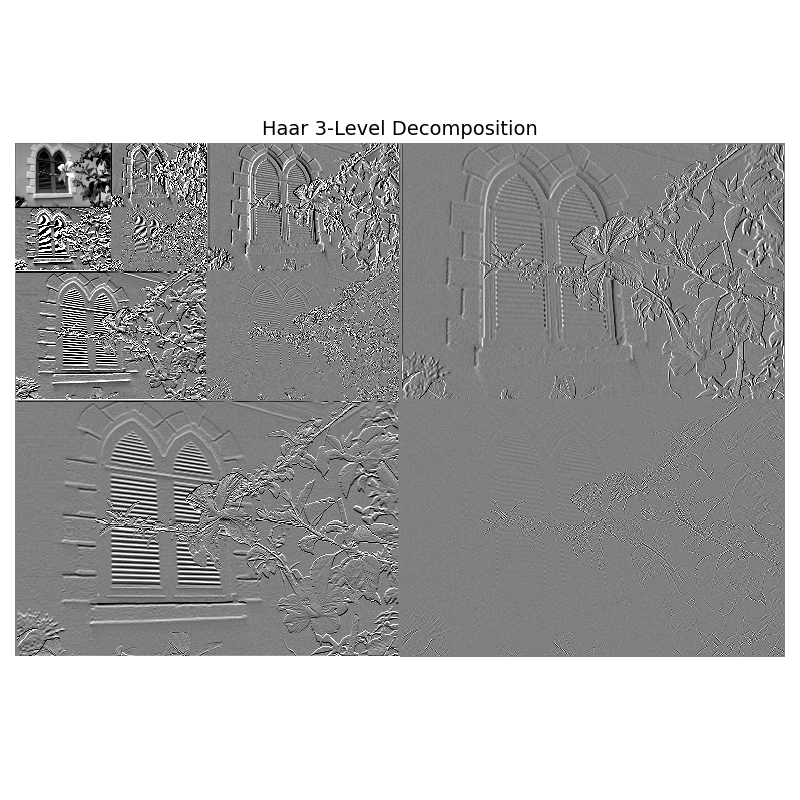
3-Level Decomposition#
wavelet3 = HaarWavelets(scales=3)
coeffs3 = pyramid_with_quadrant_embedding_levels(wavelet3, image_gray, 3)
plt.figure(figsize=(8, 8))
imshow_matched_LL(coeffs3, levels=3, orig_image=image_gray,
detail_pct=95, ll_low=5, ll_high=99,
title="Haar 3-Level Decomposition")
plt.tight_layout()
plt.show()
Total running time of the script: (0 minutes 3.555 seconds)
